Reproducible Neuroimaging: Principles and Actions
ReproNim’s principles of reproducible neuroimaging
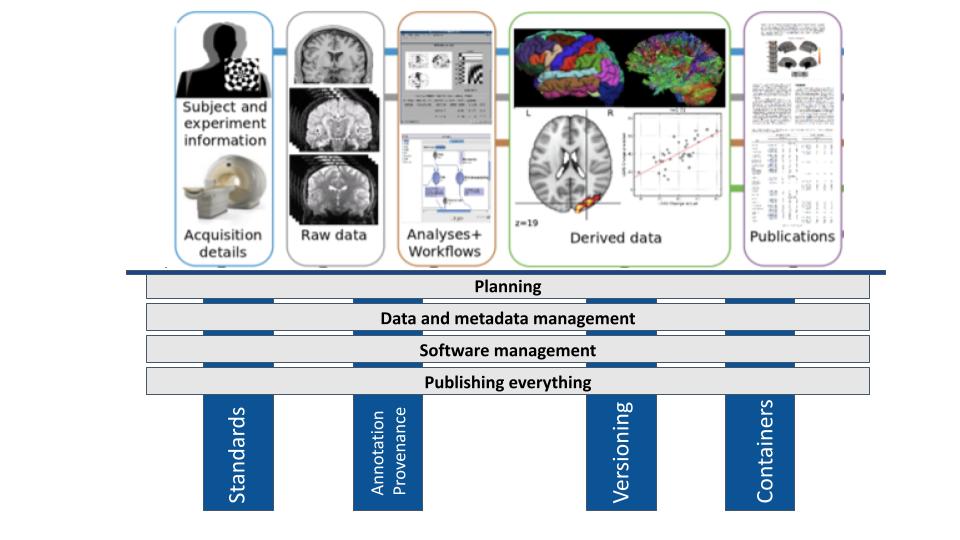
- Study planning
- Implement good science basics (power analysis, statistical consults, etc.).
- Use pre-existing data for planning and/or analysis.
- Adopt open consent to allow broad sharing of data.
- Pre-register your study.
- Data and metadata management
- Use standard data formats and extend them to meet your needs.
- Use version control from start to finish.
- Annotate data using standard, reproducible procedures.
- Software management
- Use released versions of open source software.
- Use version control from start to finish.
- Automate the installation of your code and its dependencies.
- Automate the execution of your data analysis.
- Annotate your code and workflows using standard, reproducible procedures.
- Use containers where reasonable.
- Publishing everything (publishing re-executable publications)
- Share plans (pre-registration).
- Share software.
- Share data.
- Make all research objects FAIR.
ReproNim’s four core actions
As indicated by the blue highlights in the figure below, four core actions are key to implementing the above principles.
Use of standards
Using standard data formats and extending them to meet specific research needs is important for data and metadata management (Principle 2) in reproducible neuroimaging.
Annotation and provenance
Annotating data using standard, reproducible procedures ensures clarity and transparency in data management (Principle 2). Provenance refers to the origin and history of data and processes, enabling researchers to track how data was generated, modified, and analyzed (Principles 2, 3, and 4). This is essential for understanding the context of data and ensuring reproducibility.
Implementation of version control
Version control is crucial for both data and software management. It allows researchers to track changes over time, revert to previous versions if necessary, and collaborate effectively.
For data, version control helps manage different versions of datasets and track modifications made during processing and analysis (Principle 2).
For software, version control helps track code changes, manage different versions of analysis scripts, and ensure that the correct version of the code is used for each analysis (Principle 3).
And even publications can be versioned (Principle 4).
Use of containers
Containers provide a portable and self-contained environment for running software, ensuring that the analysis can be executed consistently across different computing environments (Principle 3). Containers encapsulate all of the software dependencies needed to run an analysis, making it easier to share software (Principle 4) and reproduce results.